-
-
Site-specific mapping of cysteine redox forms -
Developing Computational Tools To Empower Chemoproteomics
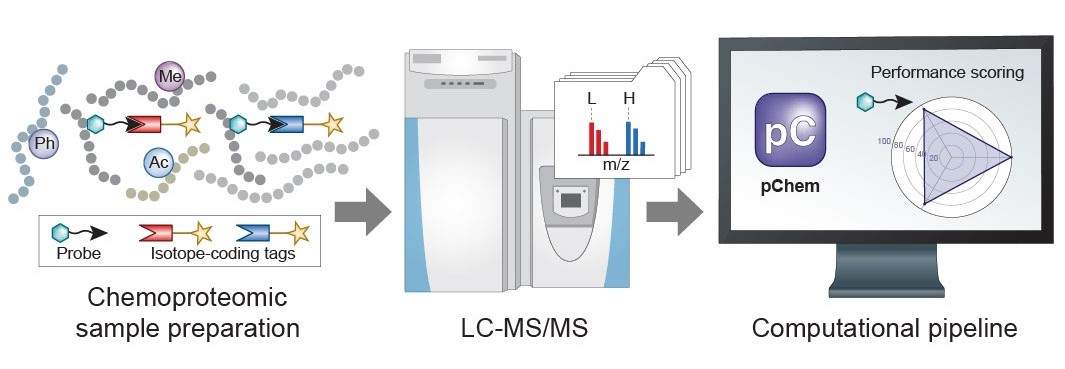
Chemoproteomics has emerged as a key technology to expand the functional space in complex proteomes for probing fundamental biology and for discovering new small-molecule-based therapies. However, developing an 'ideal' probe for chemoproteomic applications is challenging. We recently report a modification-centric computational tool termed pChem (Nat Chem Biol 2022) to provide a streamlined pipeline for unbiased performance assessment of chemoproteomic probes, thereby facilitating their development. In addition, a highly flexible nature of pChem enables its adaptation to many other interesting pursuits, such as profiling isotope-labeled RNA-protein interactions, assessing the proteome-wide reactivity of isotopically coded cross-linkers, characterizing unforeseen drug-protein adducts, discovering novel endogenous PTMs in complex proteomes, and so on.
Profiling of the Cysteine Redoxome by Chemoproteomics
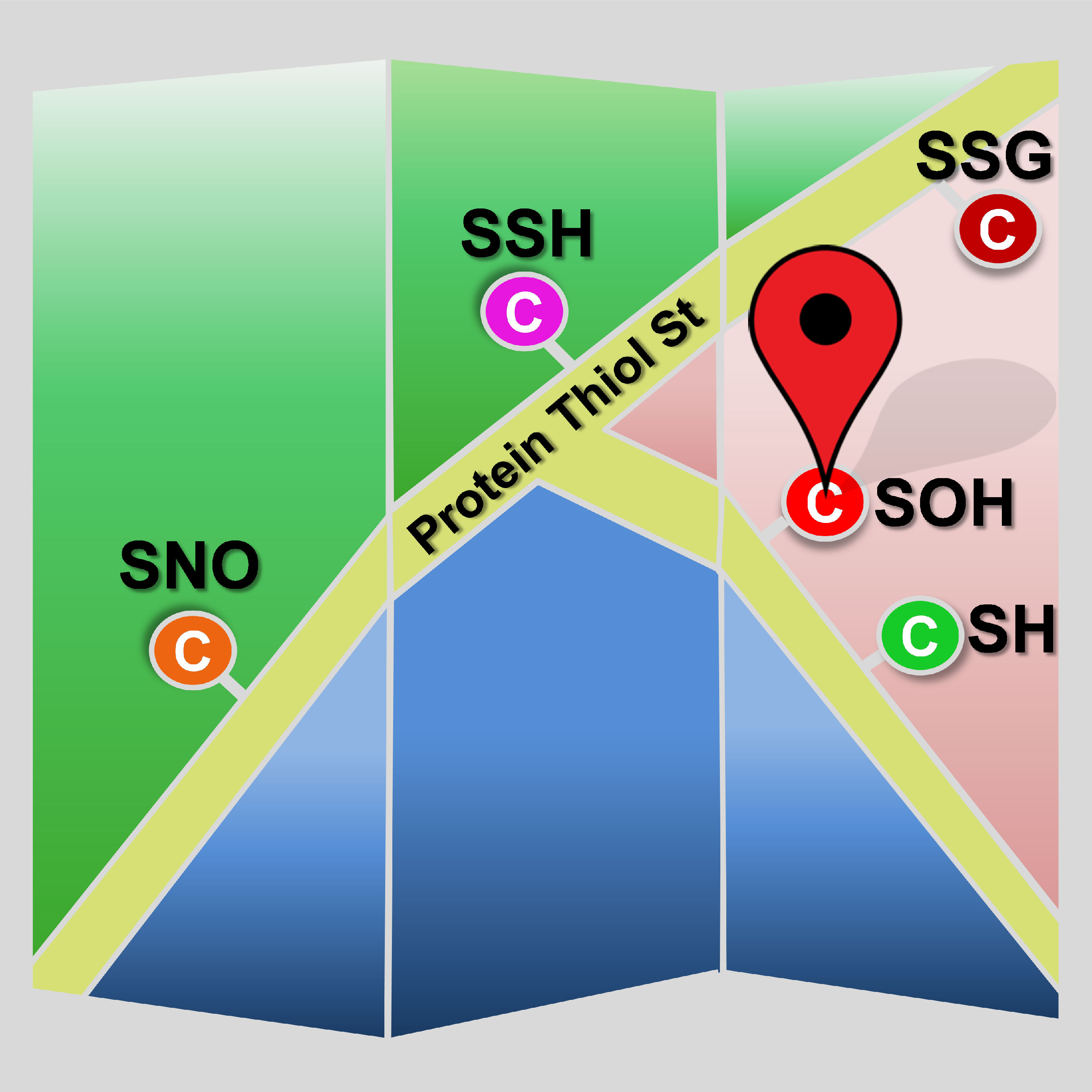
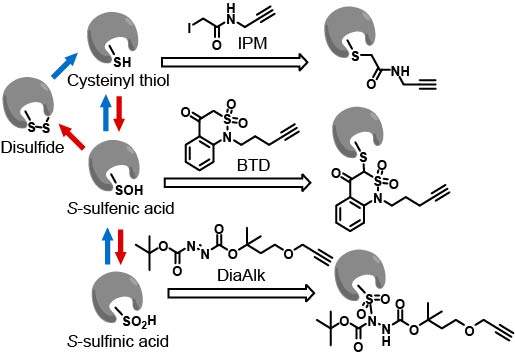
Cysteine redox modifications are well-controlled, site-specific cellular events, which play important roles in regulating many biological processes. We have unbiasedly evaluated many redox form-specific chemistries with pChem, enabling the development of a series of chemoproteomic methods to directly, site-specifically map and quantify cysteine redoxforms, including cysteinyl thiol (-SH, Nat Protoc 2020) , sulfenic acid (-SOH, sulfenylation, Nat Chem 2021), sulfinic acid (-SO2H, sulfinylation Nat Chem Biol 2018), and persulfide (-SSH, persulfidation or sulfhydration, Antioxid Redox Signal 2020). Our platform has been successfully applied to multicellular animals, such as plants (Nat Plants, 2022; Proc Natl Acad Sci U S A 2019), worms (Nat Commun 2021), flies (Proc Natl Acad Sci U S A 2018), mice (Nat Cell Biol 2019) and rats (Redox Biol 2021) for identifying proteome-wide alterations in reactive cysteines upon oxidative stress or genetically regulated redox perturbation, providing several valuable resources for uncovering the mechanisms of redox-modulated control in these model organisms. These data sets not only provide mechanistic support for prioritizing functional redox sites, but also offer novel redox mechanisms.
Expanding the Druggable Space in the Cysteine Redoxome
Covalent drugs have been successfully applied to key oncogenic driver proteins, including EGFR and mutant KRAS, historically considered to be ‘undruggable’. Despite encouraging results in the clinic, much of the human proteome still lacks defined small-molecule ligands and the number of ‘hot spots’ identified for covalent targeting remains limited. The need to expand the protein landscape that is amenable to targeting by covalent chemistry is significant and various approaches have been reported to access chemical functionality inherent to amino acid side chains. Up to now, these efforts have been directed toward cysteine. In this regard, our chemoproteomic platform also provides a potential framework for target profiling of cysteine redox form-reactive chemicals, including electrophiles for the reduced form (Cell Host Microbe 2020; Cell Chem Biol 2017; Chem Res Toxicol 2017) and nucleophiles for the oxidized form (Nat Chem Biol 2023).
Retrieving tweets.



